

To avoid under- or over-loading a gel and to ensure accurate quantification for downstream steps, scientists should measure the total protein in their samples before loading the gel. If a long shelf-life is desirable, TGX gels have a one-year shelf life. Tris-HCL gels with Tris/glycine/sodium dodecyl sulfate (SDS) buffers are cheap and easy to prepare in-house. 2 Gels with a higher percentage of polyacrylamide (10% or more) have smaller pores and can separate small proteins (250 kDa), for peptide sequencing, or mass spectrometry applications. The gel type is selected based on the molecular size of the target protein(s). For optimal separation, scientists should consider the gel type, the chemistry of the gel and the running buffer, and the total amount of protein in their sample. STEP TWO: Gel Electrophoresis with the Right MaterialsĮlectrophoresis separates the proteins contained in the cell extract by size. For example, to preserve SUMOylation, it’s best to add isopeptidase inhibitor proteins, and for ubiquitylation, ethylenediamine tetraacetic acid (EDTA) or ethylene glycol tetraacetic acid (EGTA) and iodoacetamide (IAA) or N-ethylmaleimide (NEM) should be added to the lysis buffer. For best results, the lysis buffer recipe should be adjusted according to the requirements of the target protein(s). A cocktail of protease inhibitors is added to prevent protein degradation.

Lysis releases proteases that can affect protein stability. With harsher methods, foaming should be avoided because it can decrease protein yield. Harsher methods such as sonication or French-pressing are normally required to disrupt tissues. Detergent-based or enzymatic-based methods are gentle and suitable for extracting protein from cells. 1Ī wide range of cell disruption methods exists. If the target protein yield is low, scientists should consider fractionation to enrich for low-abundance proteins. 1 For example, RIPA lysis buffer is preferred for extracting nuclear proteins. Scientists can optimize their lysis buffers by considering the cellular location of the protein. That means working with ice-cold buffers and processing tissues immediately or flash-freezing and storing tissues at -80 ☌. Moreover, contamination and protein degradation need to be minimized.Ĭlean tools, and pH-neutral water will keep contamination down, and cold materials will slow protein degradation. STEP ONE: Optimizing Lysis During Sample Preparation is Criticalĭuring sample preparation, scientists should compare lysis methods-the methods used for breaking open the cells-and optimize the lysis buffer to maximize solubility and recovery of the target protein(s). By executing best practices in these steps – sample preparation, gel electrophoresis, gel transfer, protein detection, and image analysis – researchers can expect good western blot data. This article outlines key workflow steps to help researchers perform better western blots. These results suggest that a majority of scientists find it challenging to accurately quantify their western blots and generate reliable data.
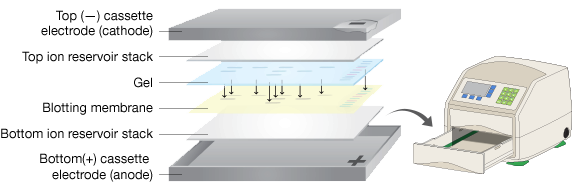
Furthermore, seven out of ten who measured variation reported that their results weren’t reproducible (%CV > 10%). When performed well, they can be accurate and cost-effective.īut, according to a Bio-Rad survey of North American scientists, nearly half reported that their western blots fail at least a quarter of the time. These forums are a place to ask questions, see answers, and share ideas with Bio Rad scientists and your peers.The goal of western blotting is to reliably detect and quantify protein expression levels. When you sign up for a class, you will automatically have access to private topic specific, online communities. Each class will include approximately 45 min of video instruction, quizzes and links to valuable western blotting resources. Western Blot Course Topics and InstructorsĬlasses are taught by Bio Rad's Field Application Specialists who have diligently compiled all the questions they have received from scientists and have designed these courses to be both comprehensive and efficient.


 0 kommentar(er)
0 kommentar(er)
